In the 2025 challenge, we leave it up to the challenge participants to determine the approach to predict brain responses (see Challenge Rules). As an example, we guide the reader through one common approach called linearizing encoding (Wu et al., 2006, Kay et al., 2008; Naselaris et al., 2011; Yamins & DiCarlo, 2016; Kriegeskorte & Douglas, 2019) where the response of each fMRI parcel is independently predicted using the multiple stimulus features provided by a computational model (linear regression is typically used to form the prediction).
We provide an example implementation of the linearizing encoding model in the development kit.
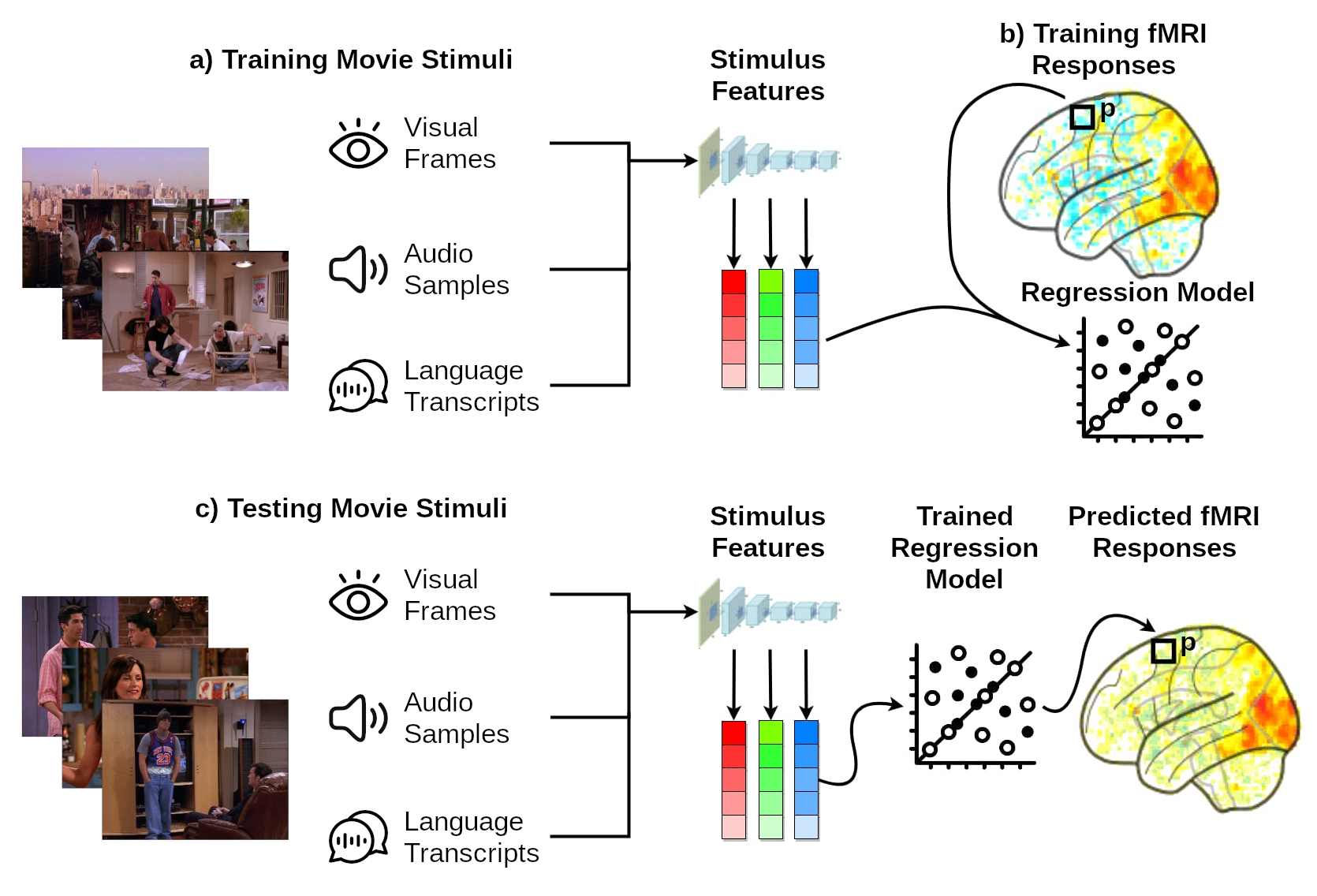
Figure 1 | Linearizing encoding approach. a) First, we use a computational model to extract the visual, audio and language features for the training movie stimuli. b) Next, we train a linear mapping (via regression) between these stimulus features and the brain parcel activity responses for the same training movie stimuli. c) Finally, we use the trained linear mapping to predict (i.e., to encode) the brain parcel responses for the testing movie stimuli.
The linearizing encoding approach has three steps:
Step 1: Feature extraction. The features for the training movie stimuli are extracted using computational models (Figure 1a).This changes the format of the data from pixels, audio samples or text (language transcripts), to model features. The features of a given model are interpreted as a potential hypothesis about the features that the brain uses to represent the stimuli.
Step 2: Model training. The stimulus features are then linearly mapped onto each brain parcel’s responses (Figure 1b). This step is needed as there is not necessarily a one-to-one mapping between stimulus features and brain parcels. Instead, each parcel’s response is hypothesized to correspond to a weighted combination of activations of multiple features.
Step 3: Model testing. The estimated linear mapping is applied on the stimulus features for the testing movie stimuli, thus predicting the corresponding fMRI responses to these stimuli (Figure 1c). The predicted fMRI test data is then compared against the ground-truth (withheld) fMRI test data. If the computational model is a suitable model of the brain, the predictions of the encoding model for the test movie stimuli will fit empirically recorded data well. In the challenge, we compare your predicted brain responses for the test movie stimuli against the corresponding empirically recorded fMRI responses. If you want to evaluate your model yourself before you submit, you can do so by dividing the training data we provide you into training and validation partitions. The development kit provides an example of how to do this.
Wu, M. C.-K., David, S. V., & Gallant, J. L. (2006). Complete Functional Characterization of Sensory Neurons by System Identification. Annual Review of Neuroscience, 29(1), 477–505. DOI: https://doi.org/10.1146/annurev.neuro.29.051605.113024
Kay, K.N., Naselaris, T., Prenger, R.J. and Gallant, J.L. (2008). Identifying natural images from human brain activity. Nature, 452(7185), pp.352-355. DOI: https://doi.org/10.1038/nature06713
Naselaris, T., Kay, K. N., Nishimoto, S., & Gallant, J. L. (2011). Encoding and decoding in fMRI. Neuroimage, 56(2), 400-410. DOI: https://doi.org/10.1016/j.neuroimage.2010.07.073
Yamins, D.L. and DiCarlo, J.J. (2016). Using goal-driven deep learning models to understand sensory cortex. Nature neuroscience, 19(3), pp.356-365. DOI: https://doi.org/10.1038/nn.4244
Kriegeskorte, N. and Douglas, P.K. (2019). Interpreting encoding and decoding models. Current opinion in neurobiology, 55, pp.167-179. DOI: https://doi.org/10.1016/j.conb.2019.04.002